Bacterial Diversity in Soil Surround Subterranean Termites-Damaged Wooden Buildings in Seonamsa Temple and Effect of the Termites on Bacterial Diversity in Humus Soil
Article information
Abstract
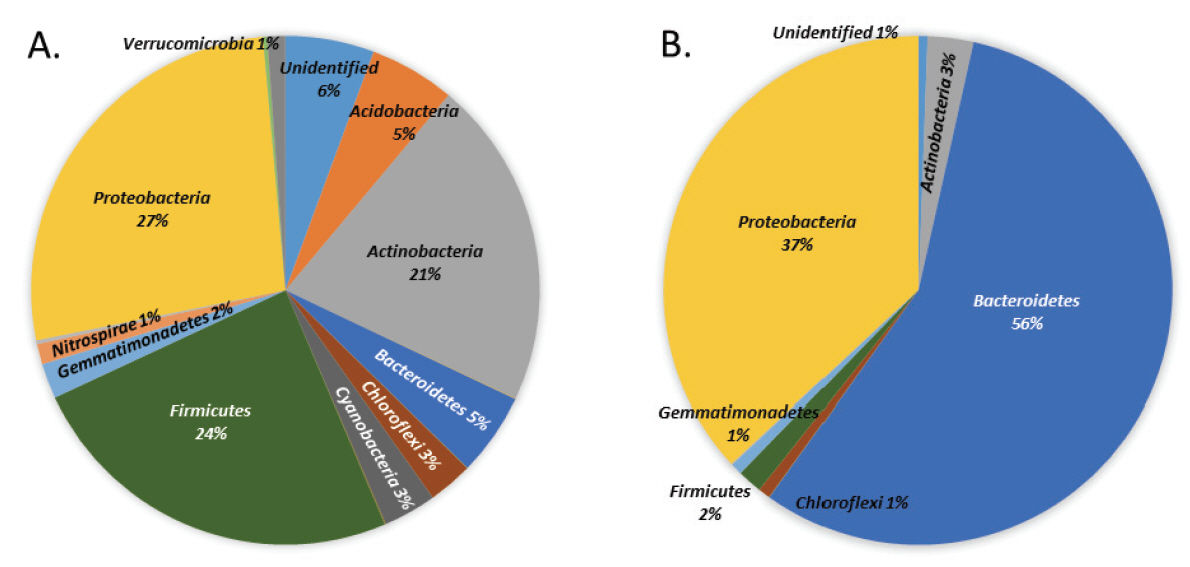
In order to determine the changes in microbial community due to termites, soil microorganisms surrounding the termites were investigated. First, bacterial communities from soil with termites collected at Seonamsa temple, Suncheon city, Korea were compared by next-generation sequencing (NGS, Illumina Miseq). The bacterial composition of soil from Daeungjeon without termites and the soil from Josadang, Palsangjeon, and Samjeon with termites were compared. Next, the bacterial composition of these soils was also compared with that of humus soil cultured with termites. A total high-quality sequences of 71,942 and 72,429 reads were identified in Seonamsa temple’s soil and humus soil, respectively. The dominant phyla in the collected Seonamsa temple’s soil were Proteobacteria (27%), Firmicutes (24%) and Actinobacteria (21%), whereas those in the humus soil were Bacteriodetes (56%) and Proteobacteria (37%). Using a two-dimensional plot to explain the principal coordinate analysis of operational taxonomic unit compositions of the soil samples, it was confirmed that the samples were divided into soil with and without termites, and it was especially confirmed that the Proteobacteria phylum was increased in humus soil with termites than in humus soil without termites.
1. INTRODUCTION
The wooden cultural buildings in Korea are almost damaged by termites. Termites mainly feed on plant and wood biomass, which are rich in cellulose and hemicellulose. The lignocellulose-degrading capability of termites is associated with the composition and diversity of their gut symbionts. Termite-related microbial communities play a major role in cellulose and lignocellulose degradation. The nest and soil around the termites are composed of partially digested food materials and faecal matter and are enriched with minerals and other organic constituents (Rossmassler et al., 2015). Thus, it provides a suitable environment for a huge diversity of microorganisms. The termite gut microbial diversity is affected by feeding habitat and surrounding environments. Termites are classified into lower and higher termites based on the presence of various flagellate or protozoan populations in their gut. High termites are more dependent on bacterial communities for digestion of cellulose because flagellates or protozoa are presented only in lower termites (Thongaram et al., 2005). As per previous microbial surveys on the gut of soil-feeding termites and their nest soil, there exist differences between the microbial communities in the termite gut and nest soil. The termite gut is dominated by the phylum Firmicutes, whereas the Actinobacteria and Proteobacteria dominate the nest and surrounding soil, followed by Bacteroidetes and Acidobacteria (Fall et al., 2007; Manjula et al., 2016).
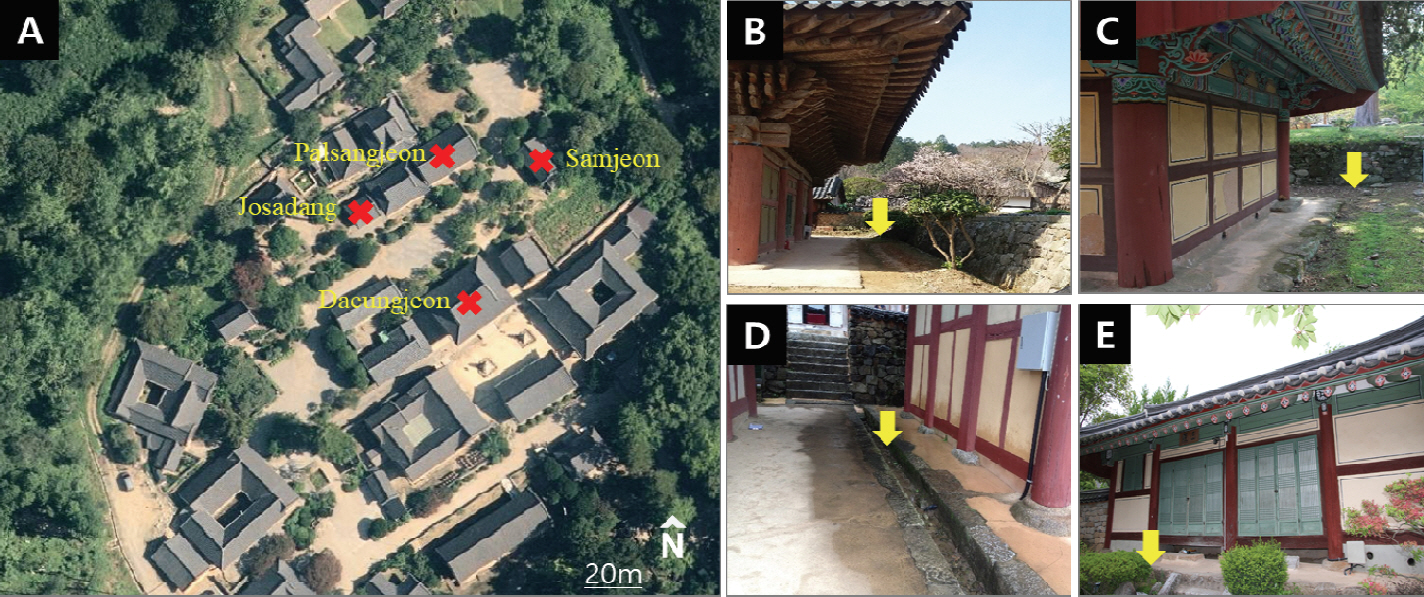
The aim of this study was to investigate the bacterial community in the soil surrounding the buildings damaged by termites and to determine whether soil bacterial compositions are affected by termites. The Seonamsa temple is located in the south province in Korea; it has been found to have many severely damaged buildings during the termite damage investigation (Kim et al., 2019; Lim et al., 2019). The result of monitoring through the replacement of detection timbers around the damaged buildings annually from 2016 to 2019, damage was observed periodically at the same location in Josadang hall (JSD), Palsangjeon hall (PSJ) and Samjeon hall (SJ). However, no termite damage was observed in the detection timbers around Daeungjeon hall (DUJ). The soils were collected around the detection timbers where damage was confirmed, and microbial diversity and community were investigated by Next Generation Sequencing (NGS) analysis. In another experiment using humus soil, changes in soil microbial community due to presence of termites were observed.
2. MATERIALS AND METHODS
2.1. Soil sampling location and humus soil test for NGS analysis
Soil samples were collected around the four buildings in Seonamsa temple (Figure 1A). From September 29, 2016 to October 31, 2019, 61 pieces of timber (5 × 5 × 30 cm) were installed at interval of 2 m around the seven buildings, checked and replaced periodically. A sampling point was selected at the location where termites were repeatedly detected in the timbers. The sampling location for each building is shown in Figure 1 (B-E). Termites were observed in the timbers installed around JSD, PSJ and SJ but not in the timbers around DUJ. When the timbers were removed, 50 g of soil samples were taken in triplicates from the Seonamsa temple at a depth of about 30 cm; these were refrigerated for sequencing analysis. For humus soil test, after putting 250 g of humus soil in 1 L culture bottles and adjusting the moisture to 70%, 200 termites (180 workers and 20 soldiers) were added and cultivated in an incubator at 25℃ for four weeks. Each group had five replicates. After four weeks, NGS analysis was performed on the humus soil without termites. The humus soil (Taeheung F&G, Korea) was purchased for gardening, and termite adaptability and microbial diversity were investigated before this test.
2.2. Next Generation Sequencing (NGS) analysis
Sequencing samples were prepared according to Illumina 16S Metagenomic sequencing library protocols. Forward primer 341F and reverse primer 805R for 16S rRNA amplification were used to amplify the V3-V4 region of the bacterial 16S rRNA. Gene amplification and sequencing were performed by Macrogen Ltd. (Seoul, Korea), and the final results were sequenced using the Illumina MiSeq platform (Illumina Inc., San Diego, CA, USA) (Caporaso et al., 2012). Operational taxonomic unit (OTU) clustering was used in de novo (CD-HIT) OTU picking method. Taxonomic assignment was conducted using the National Center for Biotechnology Information (NCBI) Database (BLAST).
3. RESULTS
3.1. Estimated richness and diversity of soil microbiota based on NGS analysis
Good’s coverage prediction for all soil samples was >99%, indicating that the number of sequences was satisfactory to reveal the bacterial diversity in each sample. The sequencing of each group was integrated, yielding 71,942 and 72,429 sequence reads from Seonamsa temple’s soil and humus soil samples, respectively. The diversity of bacterial communities in soil samples was predicted by Shannon’s index. In particular, the Shannon’s diversity index of SJ was the highest, followed by PSJ, DUJ, and JSD. In the termite test using humus soil, the Shannon’s diversity index of humus soil with termites was higher than that of humus soil without termites (Table 1).
3.2. Structural composition of soil microbiota at the phylum level
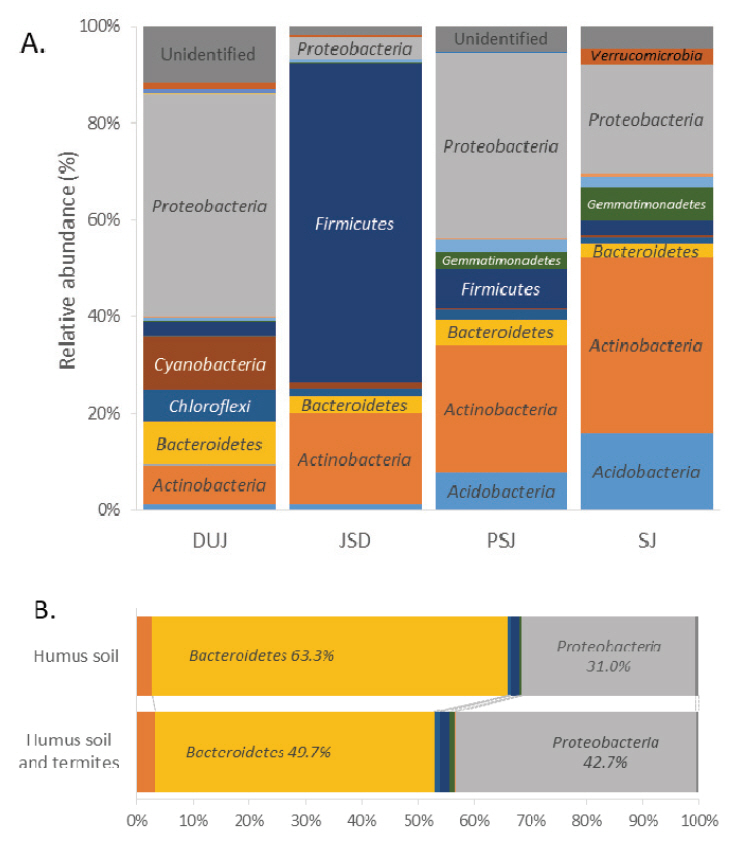
Figure 2 shows the microbial composition of soil from Seonamsa temple and humus soil samples at the phylum level. A total of 20 different phyla were observed, which represented the overall microbial diversity. On the other hand, about 10 different phyla were observed in humus soil, which represented less diversity than these of Seonamsa temple’s soil. The soil samples from Seonamsa temple were revealed dominant phyla Proteobacteria (27%), Firmicutes (24%), Actinobacteria (21%), Bacteroidetes/ Acidobacteria (5%), and other less abundant phyla such as Chloroflexi/ Cyanobacteria, Gemmatimonadetes, Verrucomicrobia and Nitrospirae (Manjula et al., 2014). The dominant phylum Proteobacteria play a major role in nitrogen fixation in termites. These results are consistent with the occurrence of nitrogen fixing Gammaproteobacteria in termite gut. The second most abundant phylum Firmicutes present in termite gut are well-known cellulose-fermenters and thrive in alkaline environments. The third dominant phylum Actinobacteria was reported to play major roles in degradation of cellulose, lignin, and chitin (Ohkuma and Kudo, 1996; Egert et al., 2003; Husseneder et al., 2010; Grieco et al., 2019). Humus soil test was revealed representative dominant phyla Bacteroidetes (56%) and Proteobacteria (36%). When the compositions of each soil samples were compared, Firmicutes and Actinobacteria predominated the area around JSD, while Proteobacteria and Actinobacteria predominated the areas around PSJ and SJ. Furthermore, Proteobacteria was the most predominant around DUJ, followed by Cyanobacteria. In humus soil experiments, Proteobacteria increased, while Bacteroidetes decreased in humus soil with termites (Figure 3).
Relative abundance (%) of Seonamsa temple and humus soil samples at the phylum level (A; Seonamsa temple, B; Humus soil).
3.3. Correlation relationship of samples using multidimensional analysis
In this study, significant differences were observed among the soil microbiotas around the four buildings in the Seonamsa temple. In a principle coordinate analysis (PCA) of soil microbiotas based on OTUs, three soil microbiotas around the JSD, PSJ and SJ buildings were clustered together, while the soil microbiota around the DUJ building was separated. The PCA produced consistent results, showing that the soil microbiotas around JSD, PSJ and SJ with termites were correlated to each other compared to those around DUJ without termites (Figure 4, red circle). Moreover, the PCA of humus soil test showed that the microbiota of humus soil with termites was closer to microbiota of the Seonamsa temple’s soil than those of humus soil without termites (Figure 4, blue circle).

Two-dimensional plot describing the PCA of OTU composition of the soil samples. This plot visualizes the clustering of soil bacterial communities based on the presence of termites. The small colored dots correspond to different soil types and OUT projections in the sample space, so the distance between the samples reflects their specificity. Soil samples (DUJ; Daeungjeon. JSD; Josadang, PSJ; Palsangjeon, SJ; Samjeon, Humus soil with and without termites).
4. DISCUSSION
This is the first study that investigated the soil bacterial communities around the termite-damaged wooden building in Korea by NGS analysis. Based on the results, it was confirmed that the soil with termite was dominated by Proteobacteria, Firmicutes and Actinobacteria phylotypes derived from the termite gut. A shift in bacterial community structure between the soil with and without termites was investigated using PCA. The results of this analysis showed that microbial structures and communities are correlated with the presence and absence of termite. The use of large soil microbial diversity data sets enables the in-depth analyses of microbial communities in these compartments derived by termites and comparisons between termite guts and various soil compartments. Although the overall microbial community in soil with termites was not established in this study, these preliminary data can be used as a method to predict termite damage.
Acknowledgements
This research was supported by the R&D project on Restoration Technology Division in National Research Institute of Cultural Heritage, the Republic of Korea.